Integration Portfolio
Metropolis-Hastings
library(ggplot2)
library(tidyverse)
library(gridExtra)
library(MASS)Random Walk Metropolis Hastings
Let’s implement a Random Walk Metropolis-Hastings algorithm with a symmetric proposal density.
rwmh <- function(start, niter, proposal, target){
# Store accepted, rejected and all samples
accepted <- matrix(NA, nrow=niter, dimnames=list(NULL, "accepted"))
rejected <- matrix(NA, nrow=niter, dimnames=list(NULL, "rejected"))
samples <- matrix(NA, nrow=niter)
# Set first sample to start and sample uniform random numbers for efficiency
z <- start
accepted[1] <- z; rejected[1] <- z; samples[1] <- z
u <- runif(niter)
# Draw candidate, calculate acceptance prob for niter-1 times
for (i in 2:niter){
cand <- proposal$sample(z)
prob <- min(1, target$density(cand)/target$density(z))
if (u[i] <= prob) {accepted[i] <- cand; z <- cand}
else {rejected[i] <- cand}
samples[i] <- z
}
result <- list("samples"=samples, "accepted"=accepted, "rejected"=rejected,
"proposal"=proposal, "target"=target)
return(result)
}Plotting Function
The following function takes the output of the Random-Walk Metropolis-Hastings algorithm and it produces two plots:
- A density of the target distribution and a histogram of the samples generated by the algorithm.
- A trace plot showing the candidates that have been proposed or accepted.
histogram_and_trace_plot <- function(out){
# Create dataframe for target line plot and for samples histogram
x_values <- seq(from=out$target$range[1], to=out$target$range[2], length.out=100)
lineplot <- data.frame(x=x_values, y=out$target$density(x_values))
histogram <- data.frame(x=out$samples)
# Line plot + histogram
bindwidth <- (out$target$range[2]-out$target$range[1]) / 50
p <- ggplot() +
geom_histogram(data=histogram, aes(x=x, stat(density)), binwidth = bindwidth,
alpha=0.5, fill="turquoise1", color="turquoise4") +
geom_line(data=lineplot, aes(x=x, y=y), size=1) +
ggtitle(paste("Target: ", out$target$name, " Proposal: ", out$proposal$name)) +
theme_minimal() +
theme(plot.title=element_text(hjust=0.5, size=15))
# Trace plot color-coded by acceptance/rejection
pair <- data.frame(cbind(out$accepted, out$rejected)) %>%
transmute(sample = coalesce(.$accepted, .$rejected),
flag = as.factor((!is.na(.$accepted))),
iteration = row_number())
q <- ggplot(data=pair) +
geom_point(aes(x=iteration, y=sample, color=flag), alpha=0.5) +
labs(color="Legend", size=15) +
scale_color_hue(labels=c("rejected", "accepted")) +
ggtitle("Trace Plot") +
theme_minimal() +
theme(plot.title=element_text(hjust=0.5, size=15))
grid.arrange(p, q, ncol=1)
}Generate Functions and Data
Below we specify some density functions and some functions for sampling from specific densities.
# Targets
gamma_density <- function(x) dgamma(x, shape=2.0, scale=2.0)
beta_density <- function(x) dbeta(x, shape1=2.0, shape2=5.0)
chisq_density <- function(x) dchisq(x, df=3.0)
normal_density <- function(x) dnorm(x, mean=3)
# Proposals
normal_sample <- function(x) rnorm(1, mean=x)
beta_sample <- function(x) rbeta(1, shape1=2.0, shape2=5.0)
gamma_sample <- function(x) rgamma(1, shape=2.0, scale=2.0)
exponential_sample <- function(x) rexp(1)
# named lists
gamma <- list("density"=gamma_density, "sample"=gamma_sample,
"name"="Gamma", "range"=c(0.0, 20.0))
beta <- list("density"=beta_density, "sample"=beta_sample,
"name"="Beta", "range"=c(0.0, 1.0))
normal <- list("density"=normal_density, "sample"=normal_sample, "name"="Normal")
chisq <- list("density"=chisq_density, "sample"=NULL,
"name"="Chi-Squared", "range"=c(0.0, 20.0))
exponential <- list("density"=dexp, "sample"=exponential_sample,
"name"="Exponential", "range"=c(0.0, 6.0))Results
In the following plots we can see that the Random-Walk Metropolis-Hastings algorithm is indeed sampling correcly.
histogram_and_trace_plot(rwmh(0.5, 100000, normal, beta))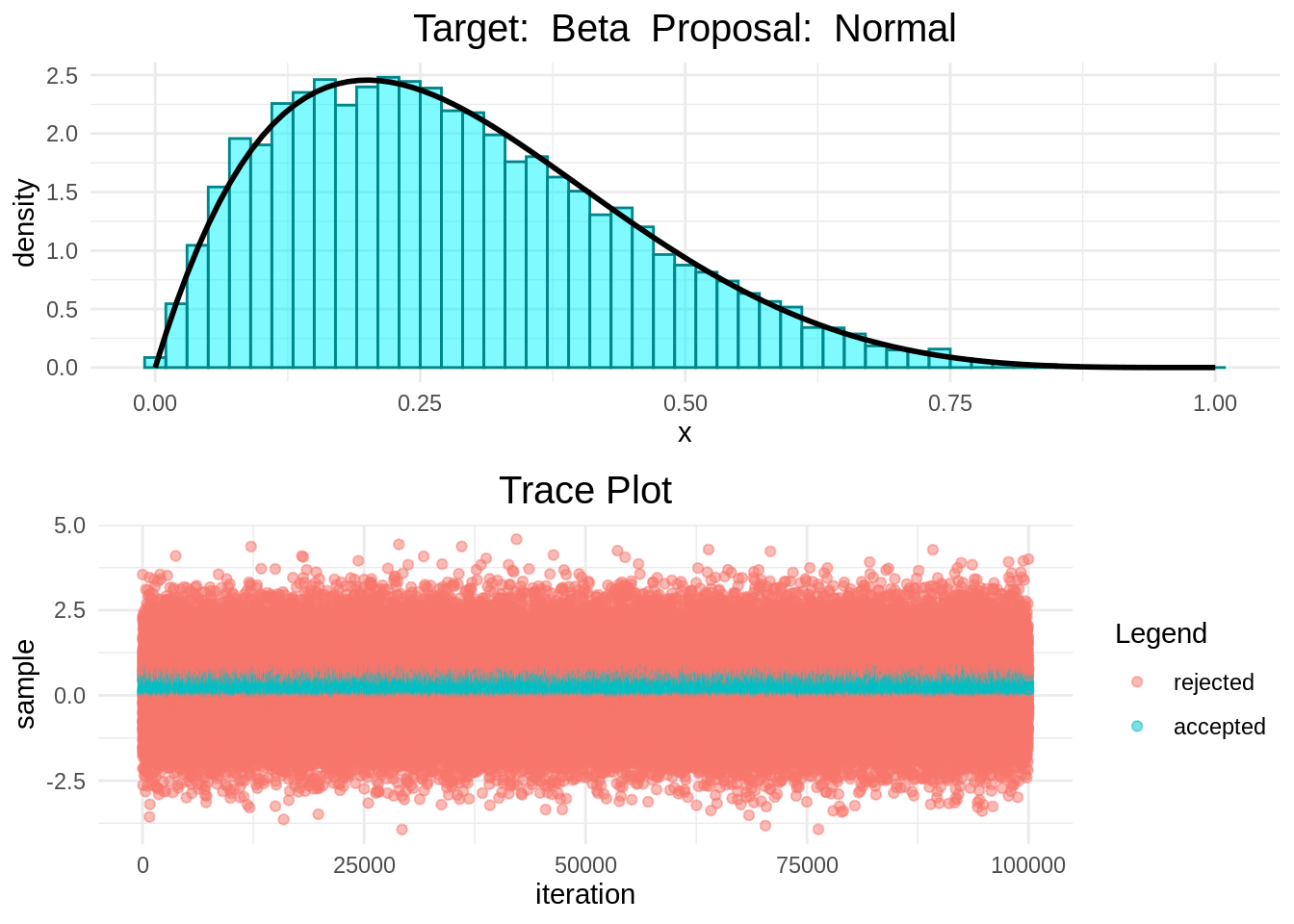
histogram_and_trace_plot(rwmh(15, 100000, normal, gamma))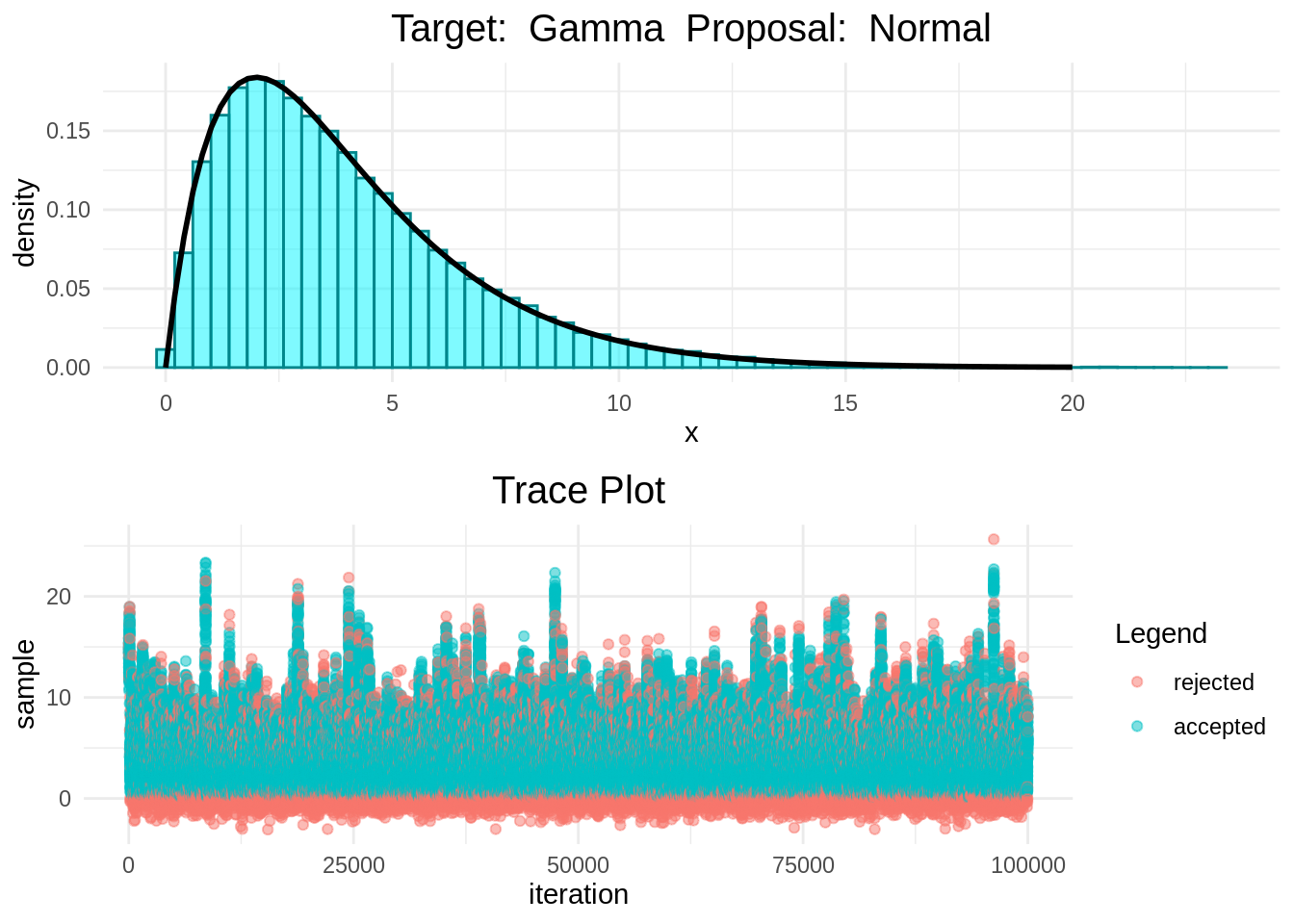
histogram_and_trace_plot(rwmh(6, 100000, normal, exponential))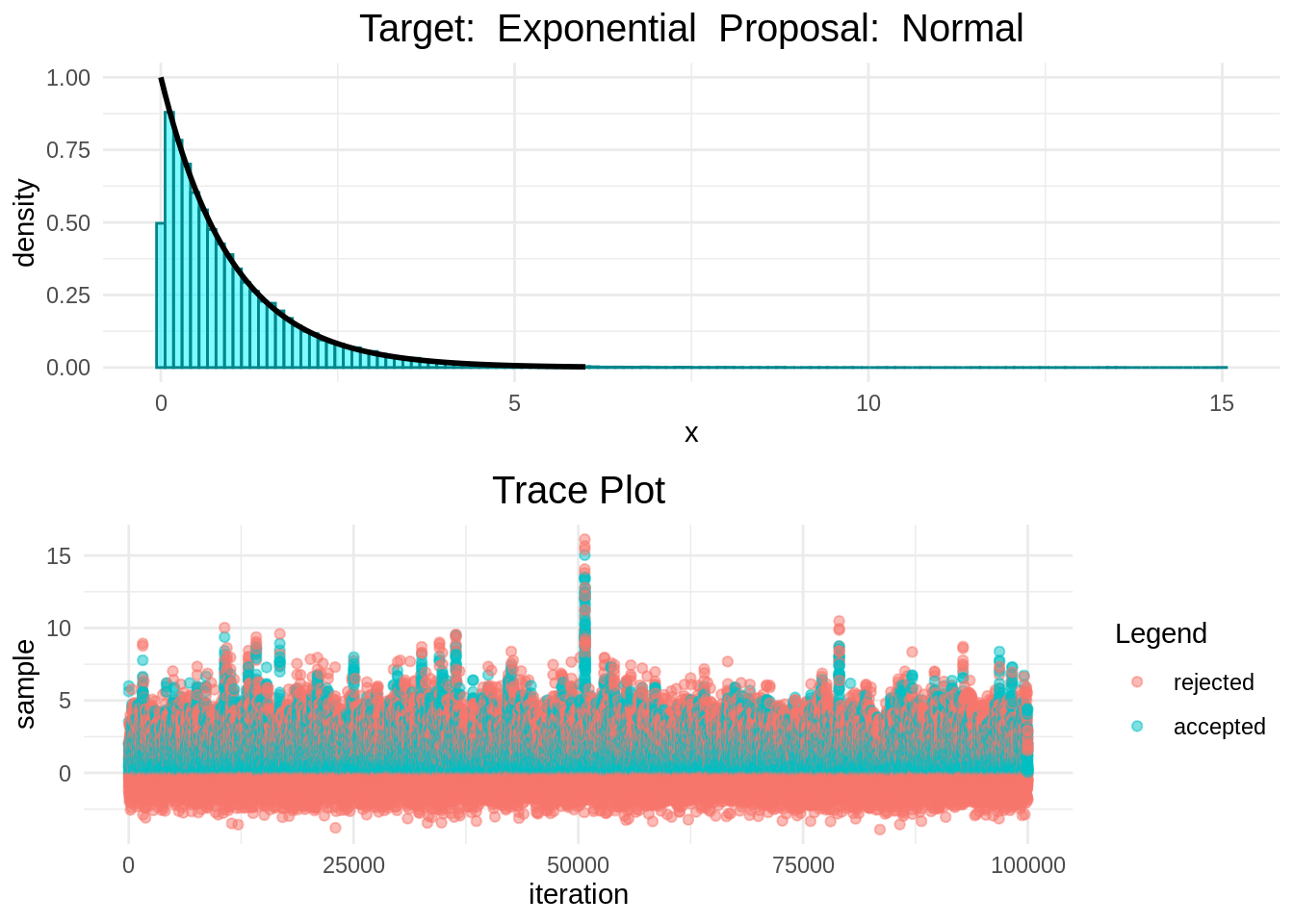
histogram_and_trace_plot(rwmh(15, 100000, normal, chisq))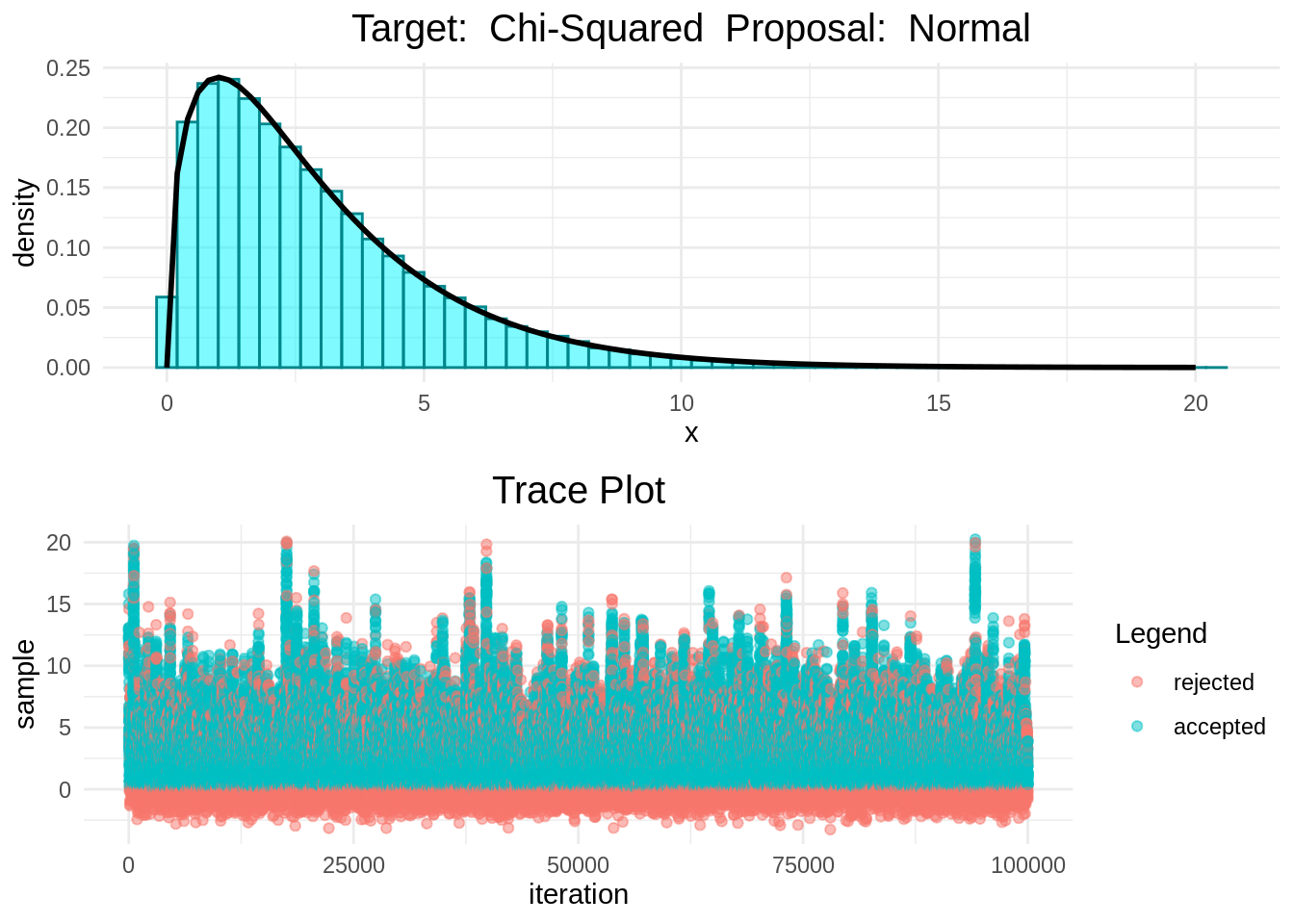
Logistic Regression Setup
Recall from the Optimization portfolio we worked with Logistic Regression.
Using an isotropic Gaussian prior on our parameters the log posterior looks like this
lnp(β∣y)∝−σ2βn∑i=1ln(1+exp((1−2yi)x⊤iβ))−12β⊤β
Now notice that this distribution is on the β, which means that our samples are samples of parameters.
Data Generation
Create data from two multivariate normal distributions.
n1 <- 100
n2 <- 100
m1 <- c(6, 6)
m2 <- c(-1, 1)
s1 <- matrix(c(1, 0, 0, 10), nrow=2, ncol=2)
s2 <- matrix(c(1, 0, 0, 10), nrow=2, ncol=2)
generate_binary_data <- function(n1, n2, m1, s1, m2, s2){
# x1, x2 and y for both classes (both 0,1 and -1,1 will be created for convenience)
class1 <- mvrnorm(n1, m1, s1)
class2 <- mvrnorm(n2, m2, s2)
y <- c(rep(0, n1), rep(1, n2)) # {0 , 1}
y2 <- c(rep(-1, n1), rep(1, n2)) # {-1, 1}
# Generate dataframe
data <- data.frame(rbind(class1, class2), y, y2)
return(data)
}
# Generate reproducible data
set.seed(123)
data <- generate_binary_data(n1, n2, m1, s1, m2, s2)
X <- data %>% dplyr::select(-y, -y2) %>% as.matrix
y <- data %>% dplyr::select(y) %>% as.matrix
# need to add another colum to x to account for the bias
X <- cbind(1, X)Multivariate Random-Walk Metropolis-Hastings Algorithm on Log Posterior
Below is a multivariate version of the Random-Walk Metropolis-Hastings algorithm defined above. This version has also been improved with the following observations:
- At the beginning samples are gonna be very much dependent on the value of
start. For this reason we introduce a number ofburniniterations that are going to be thrown away. Burn-in essentially represents the “warm up” that we want to have for our algorithm. - To remove independence we can retain every mth sample. To do this, we introduce a new parameter called
thinningthat decidesm. - We generate all uniform samples at the start and then access them when needed.
- We use linearity of the normal distribution and thus sample
nitersamples from a multivariate normal distribution and then sum them to the current value. - We also change the decision rule in a logarithm form as follows
α=min{1,p(candidate)p(current)}=min{e0,eln(p(candidate)−ln(p(current))}=min{0,ln(p(candidate)−ln(p(current))}so that sampling u∼U(0,1) and the accepting if u≤α is equivalent to sampling u∼U(0,1) and then accepting if log(u)≤ln(p(candidate)−ln(p(current))because for u∈[0,1] we have log(u)∈(−∞,0].
- Finally, we keep track of the evaluations of the log target and recycle them. This is helpful when evaluation of the log likelihood is expensive.
rwmh_multivariate_log <- function(start, niter, logtarget, vcov, thinning, burnin){
# Set current z to the initial point and calculate its log target to save computations
z <- start # It's a column vector
pz <- logtarget(start)
# create vector deciding iterations where we record the samples
store <- seq(from=(1+burnin), to=niter, by=thinning)
# Generate matrix containing samples. Initialize with the starting value
samples <- matrix(0, nrow=length(store), ncol=nrow(start))
samples[1, ] <- start
# Generate uniform random numbers in advance, to save computation. Take logarithm
log_u <- log(runif(niter))
# Proposal is a multivariate standard normal distribution. Generate samples and
# later on use linearity property of Gaussian distribution
vcov <- diag(nrow(start)) %*% vcov
normal_shift <- mvrnorm(n=niter, mu=c(0,0,0), Sigma=vcov)
for (i in 2:niter){
# Sample a candidate
candidate <- z + normal_shift[i, ]
# calculate log target of candidate and store it in case it gets accepted
p_candidate <- logtarget(candidate)
# use decision rule explained in blog posts
if (log_u[i] <= p_candidate - pz){
# Accept!
z <- candidate
pz <- p_candidate
}
# Finally add the sample to our matrix of samples
if (i %in% store) samples[which(store==i), ] <- z
}
return(samples)
}Define Function Proportional to Log Posterior and Run RWMH
# Up to normalization constant
log_posterior_unnormalized <- function(beta){
log_prior <- -0.5*sum(beta^2)
log_likelihood <- -sum(log(1 + exp((1 - 2*y) * (X %*% beta))))
return(log_prior + log_likelihood)
}
# To start the algorithm more efficiently, start from MAP estimate
optim_results <- optim(c(0,0,0),
function(x) - log_posterior_unnormalized(x),
method="BFGS", hessian=TRUE)
start <- matrix(optim_results$par)
# Use inverse of approximate hessian matrix as vcov of normal proposal
vcov <- solve(optim_results$hessian)
# Run algorithm without burnin, with no thinning
niter <- 100000
thinning <- 1
burnin <- 0
samples <- rwmh_multivariate_log(start, niter, log_posterior_unnormalized,
vcov, thinning, burnin)Trace Plots
samplesdf <- data.frame(samples) %>% mutate(iterations=row_number())
trace1 <- ggplot(data=samplesdf, aes(x=iterations, y=X1)) + geom_line()
trace2 <- ggplot(data=samplesdf, aes(x=iterations, y=X2)) + geom_line()
trace3 <- ggplot(data=samplesdf, aes(x=iterations, y=X3)) + geom_line()
grid.arrange(trace1, trace2, trace3, ncol=1)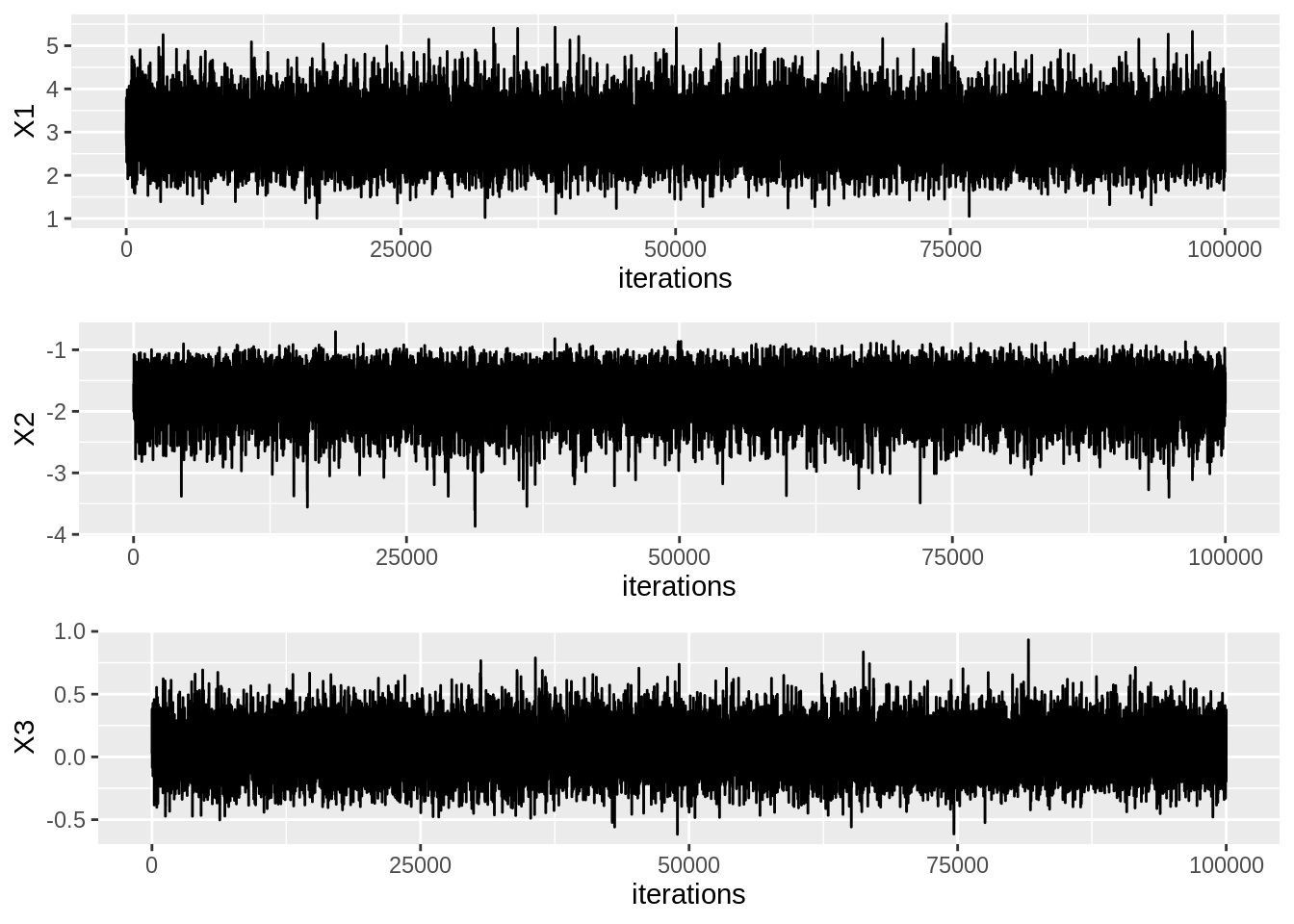
Histograms of Samples
hist1 <- ggplot(data=samplesdf, aes(x=X1, stat(density))) +
geom_histogram(binwidth=0.05, alpha=0.5, fill="turquoise1", color="turquoise4")
hist2 <- ggplot(data=samplesdf, aes(x=X2, stat(density))) +
geom_histogram(binwidth=0.05, alpha=0.5, fill="turquoise1", color="turquoise4")
hist3 <- ggplot(data=samplesdf, aes(x=X3, stat(density))) +
geom_histogram(binwidth=0.05, alpha=0.5, fill="turquoise1", color="turquoise4")
grid.arrange(hist1, hist2, hist3, ncol=1)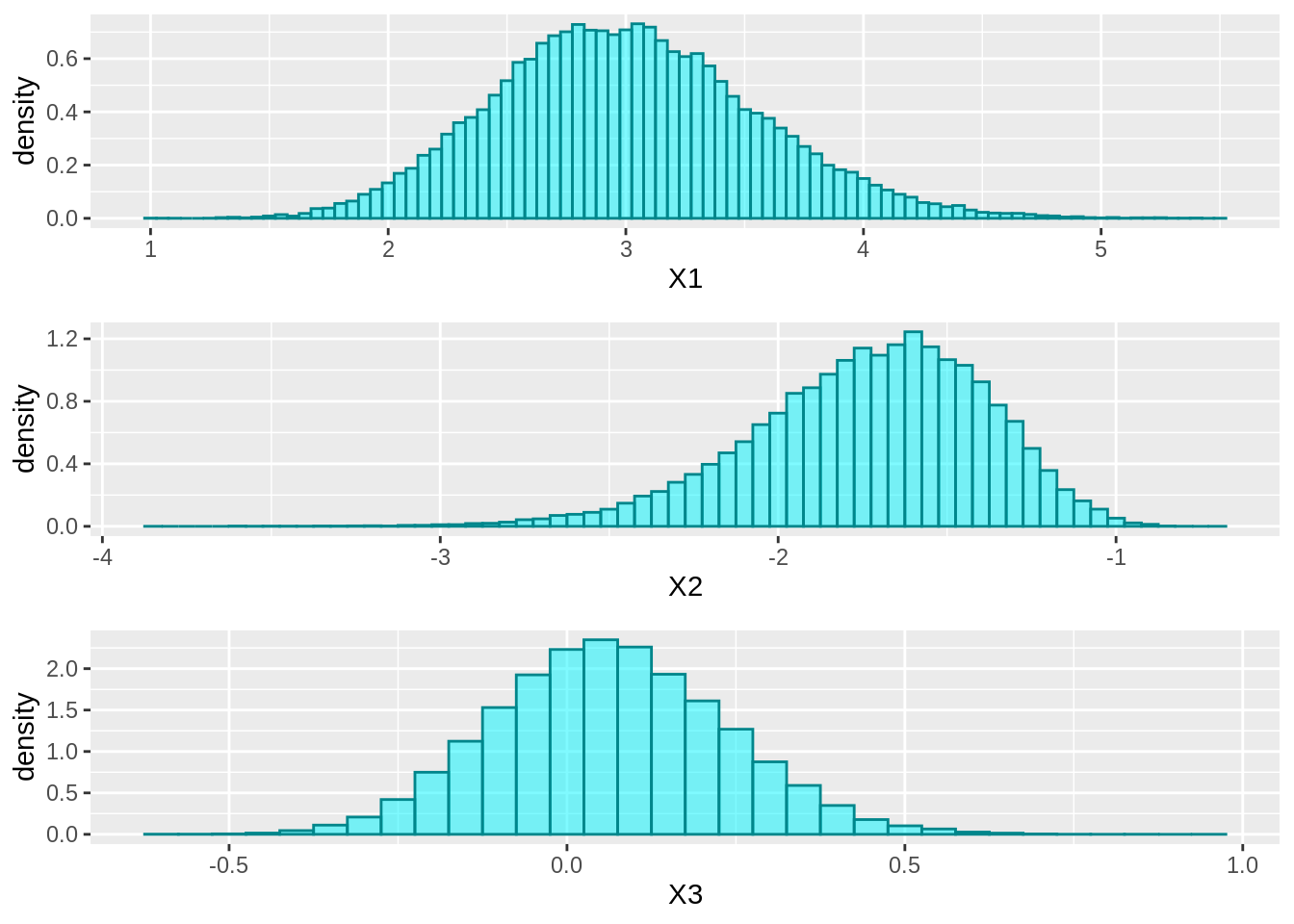
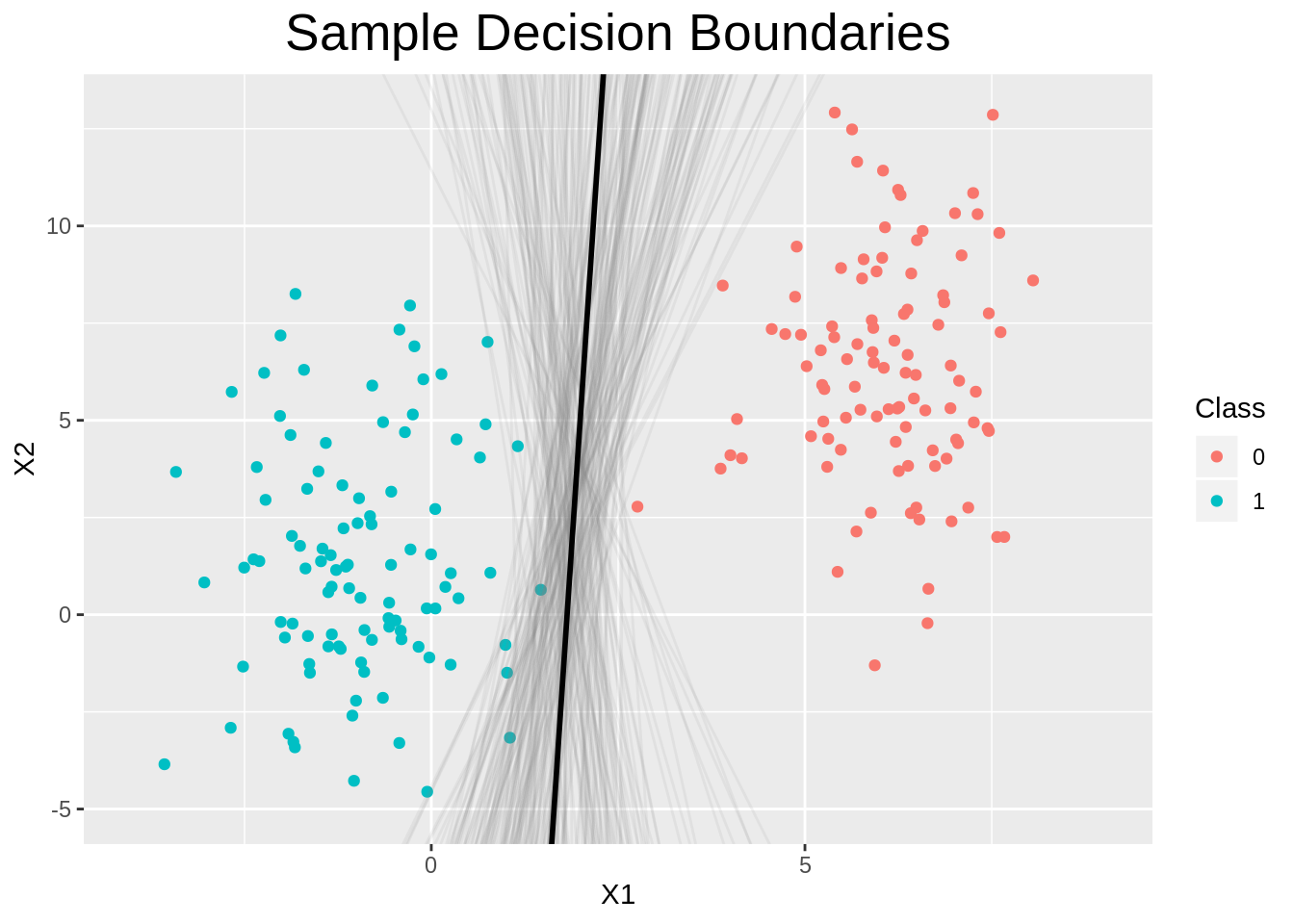
Plot decision boundary for some samples
The formula for the decision boundary is given by
x2=−β1β2x1−β0β2
samples_to_select <- 200
samples_subset <- samples[sample(1:nrow(samples), samples_to_select), ]
linecoefs <- cbind(- samples_subset[, 2] / samples_subset[, 3],
- samples_subset[, 1] / samples_subset[, 3])
x2_vals <- apply(linecoefs, 1, function(row) X[, 2]*row[1] + row[2])
dfsample_lines <- data.frame(x1=X[, 2], x2_vals) %>%
gather("key", "value", -x1)
dfmap <- data.frame(x1=X[, 2],
y=(-start[2, ]/start[3, ])*X[, 2] + (-start[1, ]/start[3, ]))
ggplot() +
geom_point(data=data, aes(x=X1, y=X2, color=as_factor(y))) +
coord_cartesian(xlim=c(-4, 9), ylim=c(-5, 13)) +
geom_line(data=dfsample_lines, aes(x=x1, y=value, group=key),
alpha=0.1, color="grey50") +
geom_line(data=dfmap, aes(x=x1, y=y), color="black", size=1) +
labs(color="Class", title="Sample Decision Boundaries") +
theme(plot.title=element_text(hjust=0.5, size=20))