Functional Programming Portfolio
Mathematical Set Up
Suppose we have a training matrix X with n observations and d features
X=(x11x12…x1dx21x22…x2d⋮⋮⋱⋮xn1xn2…xnd)=(x⊤1x⊤2⋮x⊤n)∈Rn×d
- choose b rows of X and we call them centroids
x(1),…,x(b)
- calculate using some heuristic a bandwidth parameter σ2
And then, for every centroid we define a radial basis function as follows
ϕ(i)(x):=exp(−∥x−x(i)∥2σ2)∀i∈{1,…,b}for x∈Rdpseudoinverse=TRUE in the function make_predictor defined in this document.
Parameters
In the following code chunk you can set up n the number of training observations, d the dimensionality of the input (i.e. the number of features), λ the regularization coefficient, m the number of testing observations, and b dimensionality of the data after the feature transform, which in this case corresponds to the number of centroids.
n <- 1000
d <- 2
lambda <- 0.00001
m <- 200
b <- 50Training and Testing Sets
We work with synthetic data sets. For simplicity, we will generate X uniformly in an interval (which in this case is [−4,4]) and then y will be taken to be a multivariate normal of each row of X. Note that we create a factory of functions to generate multivariate normal closures, yet the same behavior would be achieved writing up a simple function. The factory of functions is defined in order to work with the functional programming paradigm.
# Explanatory variable is uniformly distributed between -4 and 4, then reshaped
X <- matrix(runif(n*d, min=-4, max=4), nrow=n, ncol=d)
# Factory of multivariate normal distributions
normal_factory <- function(mean_vector, cov_matrix){
d <- nrow(cov_matrix)
normal_pdf <- function(x){
exponent <- -mahalanobis(x, center=mean_vector, cov=cov_matrix) / 2
return(
(2*pi)^(-d/2) * det(cov_matrix)^(-1/2) * exp(exponent)
)
}
}
# Closure will be our data-generating process for training and test response.
target_normal <- normal_factory(rnorm(d), diag(d))
y <- matrix(apply(X, 1, target_normal))Similarly, create test data.
Xtest <- matrix(runif(m*d, min=-4, max=4), nrow=m, ncol=d)
ytest <- matrix(apply(Xtest, 1, target_normal))Feature Transform Implementation
Define a function that, given a training data matrix X, finds the distance matrix containing all the pairwise distances squared, and then return the median of such distances. Essentially, it returns the badwidth squared σ2.
compute_sigmasq <- function(X){
# Compute distance matrix expanding the norm. Return its median
Xcross <- tcrossprod(X)
Xnorms <- matrix(diag(Xcross), nrow(X), nrow(X), byrow=TRUE)
return(median(Xnorms - 2*Xcross + t(Xnorms)))
}Define a function that gets b random samples from the rows of an input matrix X, these will be the centroids.
get_centroids <- function(X, n_centroids){
# Find indeces of centroids
idx <- sample(1:nrow(X), n_centroids)
return(X[idx, ])
}Construct a factory of Radial Basis Functions. The factory constructs an RBF, given a centroid and a σ2.
rbf <- function(centroid, sigmasq){
rbfdot <- function(x){
return(
exp(-sum((x - centroid)^2) / sigmasq)
)
}
return(rbfdot)
}Finally, we can put all of these functions together to compute Φ and eventually make predictions.
compute_phiX <- function(X, centroids, sigmasq){
# Create a list of rbfdot functions and apply each of them to every row of X
phiX <- mapply(
function(f) apply(X, 1, f),
apply(centroids, 1, rbf, sigmasq=sigmasq)
)
# Reshape phiX correctly
phiX <- matrix(phiX, nrow(X), nrow(centroids))
# Recall we need the first column to contain 1s for the bias
return(cbind(1, phiX)) # this is a (n, d+1) matrix
}Prediction
We’re now ready to define a factory of functions that returns prediction functions.
library(corpcor)
make_predictor <- function(X, y, lambda, n_centroids, pseudoinverse=FALSE){
# Randomly sample centoids
centroids <- get_centroids(X, n_centroids)
sigmasq <- compute_sigmasq(X)
# Get transformed data matrix
phiX <- compute_phiX(X, centroids, sigmasq)
# Find optimal parameters
if (pseudoinverse){
# This method works best cause it does automatic regularization with SVD
w <- pseudoinverse(phiX) %*% y
} else {
w <- solve(t(phiX)%*%phiX + lambda*diag(n_centroids+1), t(phiX) %*% y)
}
# Construct predictor closure for a new batch of testing data Xtest
predictor <- function(Xtest){
# Need to transform test data into a phi matrix
phi_Xtest <- compute_phiX(Xtest, centroids, sigmasq)
return(phi_Xtest %*% w)
}
return(predictor)
}We can test its effectiveness now. Notice that in general we might need a lot of centroids to make this work.
Construct prediction functions. One with regularization, other with pseudoinv
predict <- make_predictor(X, y, lambda, n_centroids = b)
pseudo_predict <- make_predictor(X, y, lambda, n_centroids = b, pseudoinverse = TRUE)
Get predictions
yhat <- predict(Xtest)
yhat_pseudo <- pseudo_predict(Xtest)Predictions can be assessed by plotting predicted values against the true test values.
library(ggplot2)
# Put data into a dataframe for ggplot2
df <- data.frame(real=ytest, pred=yhat, pseudo_pred=yhat_pseudo)ggplot(data=df) +
geom_point(aes(x=real, y=pred), color="darkblue") +
geom_abline(intercept=0, slope=1) +
ggtitle(paste("Predictions vs Actual with b =", b)) +
theme(
plot.title = element_text(hjust = 0.5),
axis.title = element_text(size=15, face="bold"),
axis.title.y = element_text(angle=0, vjust=0.5)
) +
xlab(expression(y)) +
ylab(expression(hat(y))) +
coord_fixed()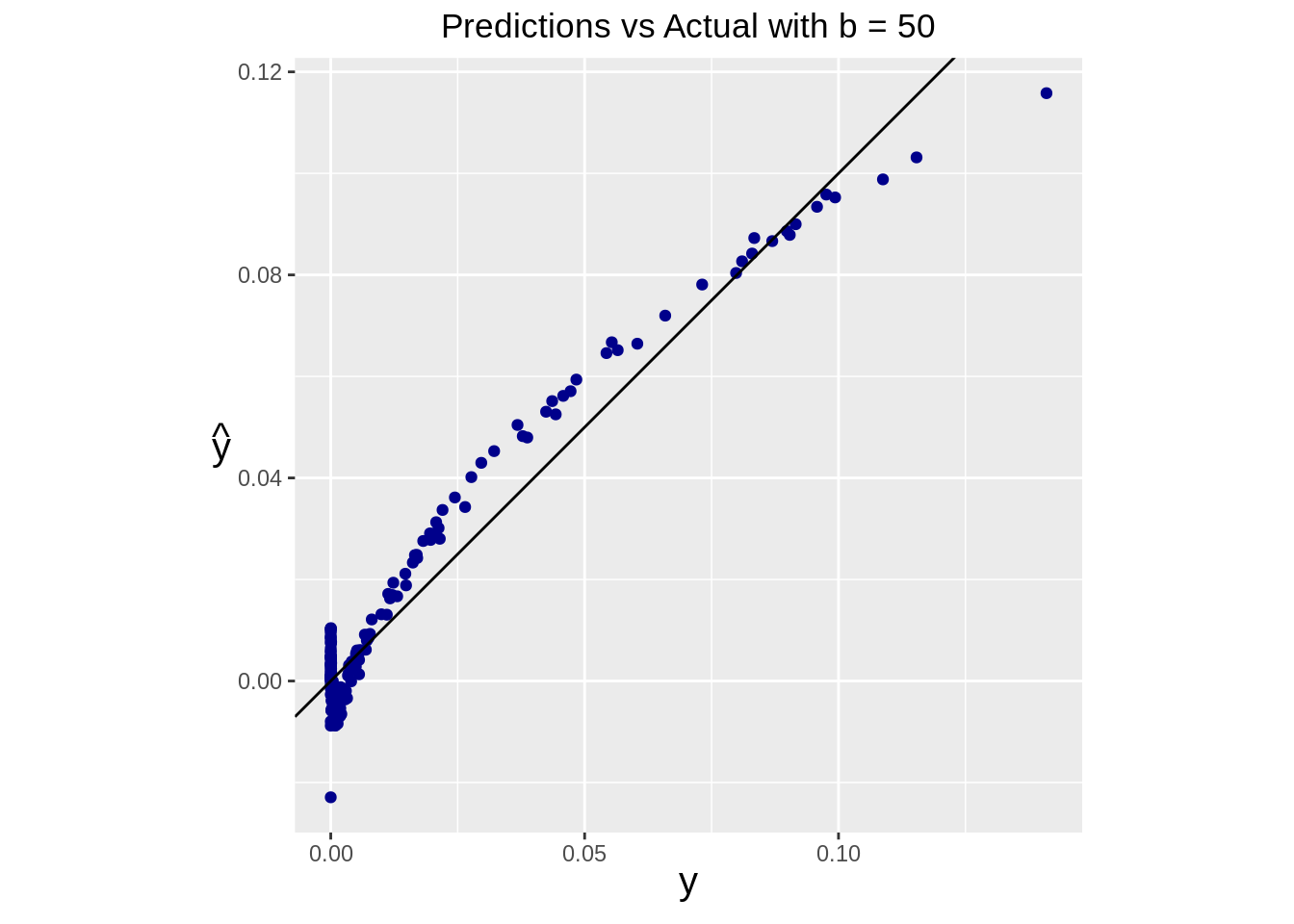
Similarly, we can see the performance of our prediction using the pseudoinverse
ggplot(data=df) +
geom_point(aes(x=real, y=pseudo_pred), color="darkblue") +
geom_abline(intercept=0, slope=1) +
ggtitle(paste("Pseudo-Predictions vs Actual with b =", b)) +
theme(
plot.title = element_text(hjust = 0.5),
axis.title = element_text(size=15, face="bold"),
axis.title.y = element_text(angle=0, vjust=0.5)
) +
xlab(expression(y)) +
ylab(expression(hat(y))) +
coord_fixed()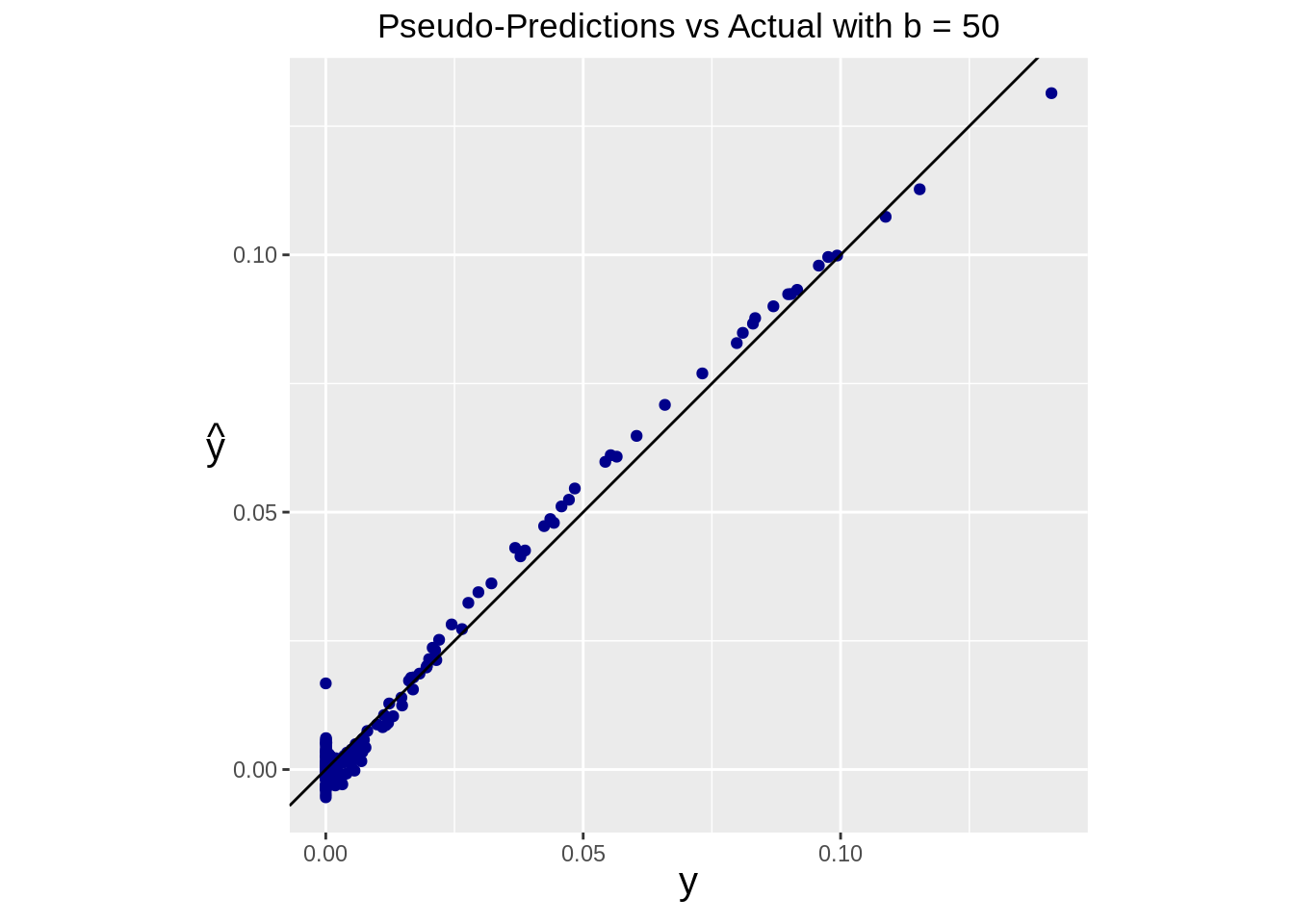
Technical Note on Calculating the Bandwidth
To vectorize this operation, we aim to create a matrix containing all the squared norms of the difference between the vectors
D=(∥x1−x1∥2∥x1−x2∥2⋯∥x1−xn∥2∥x2−x1∥2∥x2−x2∥2⋯∥x2−xn∥2⋮⋮⋱⋮∥xn−x1∥2∥xn−x2∥2⋯∥xn−xn∥2)
The key is not to notice that we can obtain the middle matrix from X with a simple operation:
XX⊤=(x⊤1x⊤2⋮x⊤n)(x1x2⋯xn)=(x⊤1x1x⊤1x2⋯x⊤1xnx⊤2x1x⊤2x2⋯x⊤2xn⋮⋮⋱⋮x⊤nx1x⊤nx2⋯x⊤nxn)
and then the first matrix can be obtained as follows
(11⋮1)n×1(x⊤1x1x⊤2x2⋯x⊤nxn)1×n=(x⊤1x1x⊤1x1⋯x⊤1x1x⊤2x2x⊤2x2⋯x⊤2x2⋮⋮⋱⋮x⊤nxnx⊤nxn⋯x⊤nxn)
